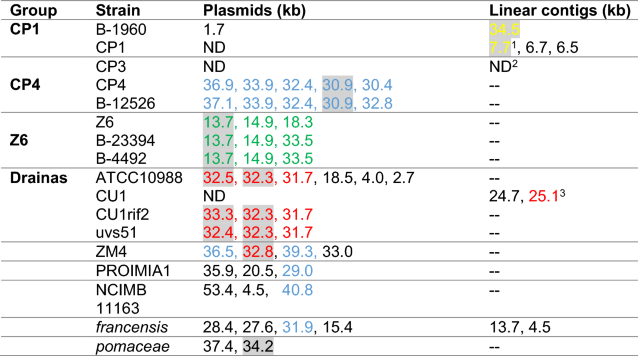
- Plasmids are named by size as determined in NCBI nucleotide database. Full names and NCBI accession numbers of all plasmids are listed in Additional file 1: Table S3. BLASTn of whole plasmids sequences was used to determine their ANI. Plasmids shared by more than one strain in the group were assigned the same colors, i.e., all blue-labeled plasmid were shared by multiple strains in the CP4 group. Homologous plasmids (> 99% ANI) are vertically aligned, i.e., the blue-labeled plasmids in the ZM4 row are homologous to the corresponding blue-labeled plasmids in the CP4 row. Plasmids with no significant homology to other plasmids are shown in black. Plasmids with annotated hsdRMS genes are highlighted (see details in text). ND-not detected
- 1hsdMS and truncated hsdR with 100% aa identity to hsdRMS on linear contig 34.5 of B-1960
- 2The CP3 chromosome could not be assembled into a circular contig; the genome could only be assembled into 22 linear contigs. Therefore, we could not distinguish between putative chromosomal and plasmid contigs.
- 399% nucleotide identity to p32.3 of ATCC10988 but no hsdRMS genes

