- Research
- Open access
- Published:
Determinants of selection in yeast evolved by genome shuffling
Biotechnology for Biofuels volume 11, Article number: 282 (2018)
Abstract
Background
Genome shuffling (GS) is a widely adopted methodology for the evolutionary engineering of desirable traits in industrially relevant microorganisms. We have previously used genome shuffling to generate a strain of Saccharomyces cerevisiae that is tolerant to the growth inhibitors found in a lignocellulosic hydrolysate. In this study, we expand on previous work by performing a population-wide genomic survey of our genome shuffling experiment and dissecting the molecular determinants of the evolved phenotype.
Results
Whole population whole-genome sequencing was used to survey mutations selected during the experiment and extract allele frequency time series. Using growth curve assays on single point mutants and backcrossed derivatives, we explored the genetic architecture of the selected phenotype and detected examples of epistasis. Our results reveal cohorts of strongly correlated mutations, suggesting prevalent genetic hitchhiking and the presence of pre-existing founder mutations. From the patterns of apparent selection and the results of direct phenotypic assays, our results identify key driver mutations and deleterious hitchhikers.
Conclusions
We use these data to propose a model of inhibitor tolerance in our GS mutants. Our results also suggest a role for compensatory evolution and epistasis in our genome shuffling experiment and illustrate the impact of historical contingency on the outcomes of evolutionary engineering.
Background
Genome shuffling (GS) is an evolutionary engineering method based on recursive recombination and selection in populations of mutants (Fig. 1). It aims to speed the rate of evolution of desired traits by exploiting sexual, parasexual or artificial recombination to promote purifying selection, positive epistasis, and the accumulation of beneficial mutations, while reducing clonal interference. It has been widely and successfully adopted for the evolutionary engineering of industrial traits in microbes [1]. GS and other evolutionary engineering methodologies are notably useful to enhance complex phenotypes for which a detailed molecular level understanding is lacking. Studies aimed at uncovering the genetic architecture of strains evolved by genome shuffling may thus contribute to an understanding of the genetic basis of complex and industrially relevant traits. Targeted approaches such as candidate gene sequencing [2] and qPCR [3, 4] have been used to uncover the genetic determinants of traits evolved by GS. System-level approaches, like array-comparative genome hybridization [5], RNAseq [5, 6], and whole genome sequencing [6,7,8], as well as proteomics methods [9,10,11] have also been used to investigate the complex genetic architecture of strains derived from GS experiments.
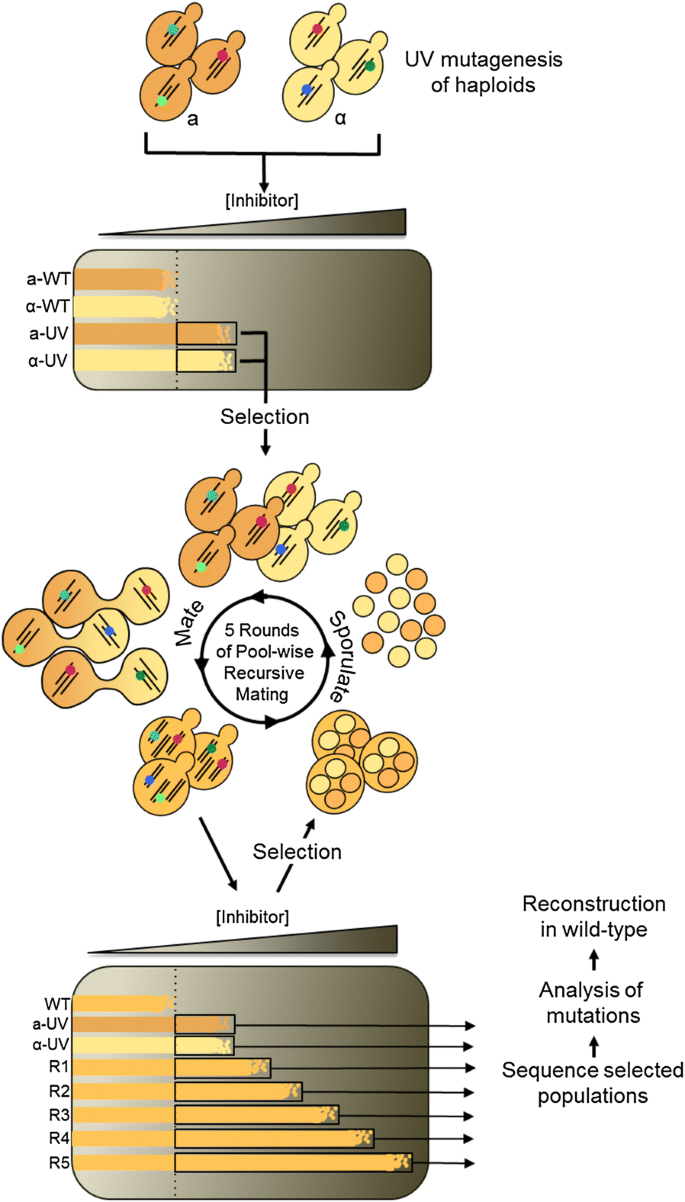
Outline of the genome shuffling experiment. Wild-type haploid cells of both mating types were UV irradiated to generate pools of haploid mutants. Mutants with tolerance to hydrolysate superior to their wild-type ancestors were selected using plates displaying a gradient of hydrolysate concentration. The pools of tolerant haploid mutants thus obtained were mated, generating diploids. Tolerant diploids were selected on gradient plates, and sporulated. Resulting haploids were mated at random, effecting the shuffling of mutations. Five cycles of diploid selection on gradient plates, sporulation and mating were performed. Genomic DNA from each round of shuffling and selection was submitted to population genome sequencing and mutation analysis
Massively parallel sequencing technologies have enabled monitoring of the appearance, frequency and fluctuation of mutant alleles in experimental evolution experiments [12]. A growing body of work on adaptive evolution takes advantage of evolve-and-resequence experiments to explore evolutionary dynamics under various sets of experimental constraints and environments [13]. Several evolutionary behaviors and dynamics predicted by evolutionary theory and their effects on adaptation have been illustrated in this way. Notable examples include clonal interference and genetic hitchhiking in asexual populations [14], the effect of sign epistasis on adaptive landscapes [15] and the mechanisms by which sexual recombination speeds adaptation [16]. Recently, whole population sequencing of evolutionary time points has been extended to the study of industrially relevant phenotypes, probing the dynamics and molecular processes affected during adaptation of S. cerevisiae to high ethanol stress [17]. This study identified specific mutations conferring increased tolerance to high ethanol and illustrated the diversity of evolutionary mechanisms involved in the adaptive response to complex stresses.
Most evolve-and-resequence studies have studied evolution from isogenic starting populations of asexually reproducing microbes [13]. These experimental setups, in which selection is applied on diversity that strictly derives from de novo mutations, have demonstrated pervasive clonal interference and widespread genetic hitchhiking [14, 18, 19]. Parallel evolution is observed in these experiments, despite high levels of molecular diversity [20,21,22]. More relevant to the context of genome shuffling, evolution of S. cerevisiae populations with sexual reproduction and high levels of initial diversity were shown to follow similarly deterministic paths while granting a marginal role for de novo mutations [23]. A similar methodology was used to compare the rate and dynamics of adaptation in the absence and presence of sexual recombination events, showing that sex accelerates the rate of adaptation by reducing clonal interference and enabling efficient purifying selection [16].
We have previously used GS to successfully evolve strains of Saccharomyces cerevisiae tolerant to spent sulfite liquor (SSL), a toxic lignocellulosic hydrolysate and byproduct of the acid bisulfite wood pulping process [24]. These mutants were characterized as highly tolerant to osmotic and oxidative stresses, organic acids and phenolic compounds. A strain, designated R57, with high inhibitor tolerance and ability to ferment hydrolysate sugars to ethanol was identified [6]. Whole-genome sequencing, RNAseq and whole population amplicon sequencing were used to probe the genetic architecture of R57. The strain differs from its parent by 21 single nucleotide changes affecting 17 genes [6].
This study aims to explore the evolutionary dynamics of genome shuffling by identifying the molecular and evolutionary determinants of selection in our experiment. To our knowledge, it is the first study to use genome sequencing of a GS population at several evolutionary time points. It completes our survey of selected mutations and retrieves allele frequency time series spanning our evolutionary engineering experiment. Inhibitor tolerance assays and genotyping by amplicon sequencing of meiotic segregants of R57 provided data for a multivariable linear model predicting the contribution of individual mutations to the hydrolysate tolerance phenotype. The phenotypic effects of single mutations reconstituted in wild type or deleted from the R57 background were also tested. Based on these results, we propose a descriptive model of the evolutionary dynamics of our GS experiment. We discuss the impact of historical contingency and compensatory evolution on the outcomes of GS, and demonstrate prevalent genetic hitchhiking. We also identify key genetic determinants of hydrolysate inhibitor tolerance.
Results
Pooled sequencing of evolving populations
Mutant populations with increased tolerance to hydrolysate inhibitors were generated by genome shuffling as described in Fig. 1 and previous publications [6, 24]. In short, two pools of random mutants were generated by UV mutagenesis of wild-type MATa and MATα haploids. Haploid mutants with tolerance above wild-type levels were selected on gradient plates, which consist in dishes of agar medium displaying increasing concentration of hydrolysate from one end to the other. MATa and MATα mutants were mated to generate diploids carrying random combinations of mutations. These diploids were selected on gradient plates to enrich for individuals with superior hydrolysate tolerance. After mating, diploids were sporulated, digesting and sonicating to disrupt asci and eliminate non-sporulated vegetative cells. Resulting haploids were mated to effect genome shuffling. In total, the mutant pool was submitted to 5 rounds of recursive mating, selection, and sporulation, generating populations with increasing tolerance to hydrolysate inhibitors. Strain R57, which displays high tolerance to hydrolysate inhibitors, was isolated from the fifth and final round of genome shuffling. Its genome differs from the CEN.PK113-7D reference sequence [25] by the 21 single nucleotide substitutions listed in Additional file 1: Table S2.
To gain insight into the genetic landscape of our populations evolved by genome shuffling, we investigated the metagenome of seven populations from six time points (Fig. 1). For sequencing, we selected time points covering the entire length of the experiment, including both populations of selected UV haploids and shuffled mutants from each of the five rounds of genome shuffling (R1–R5). Each population was re-sequenced, generating upwards of 300 million reads, for an average of 40 billion nucleotides per sample with a mean base quality score of 35.07 (Additional file 1: Table S1). The 100 nucleotide reads were aligned to the CEN.PK113-7D reference genome [25], which is one of the parental strains used in the experiment. Mean depth of coverage oscillated between 712× and 1551×, for an average of 1091×, enabling the detection of SNPs represented in < 1% of the population (Additional file 1: Table S1).
A base error model was used for calling SNPs, distinguishing genuine mutations from sequencing errors. Filtering and manual examination resulted in a list of 188 SNPs (Tab “All_mutations” in Additional file 2). A further reduced list was prepared by excluding mutations that either were synonymous or escaped detection in at least one of the six time points (Tab “Non_synonymous_non_zero” in Additional file 2). We detected the majority of SNPs previously identified in R57 [6] most of which are found at medium to high frequency in the re-sequenced populations (Fig. 2). Previous sequencing of strain R57 found mutations in genes TOF2, DOP1 and FIT3, but they could not be detected by population sequencing, implying that their frequencies fall below our detection threshold.
Evolutionary trajectories of the most prominent clusters of mutations. Mutations arose either in the MATa (left) or MATα (right) pools. On the vertical axis are the names of the mutations, giving the closest gene, coordinates relative to that gene, and the nature of the nucleotide substitution. On the horizontal axis are each of the six evolutionary timepoints (UV, R1, R2, R3, R4, R5), and the mean allele frequency change (M). Frequency of the mutant alleles is represented by shades of green. Mean allele frequency changes are represented in shades of red (M < 1, declining frequency) to blue (M > 1, increasing frequency). Hierarchical clustering of individual evolutionary trajectories is represented by dendrograms on the left. Mutations were assigned to cohorts of mutations (a1-5, α1-3) on the basis of this clustering. Mutations present in highly tolerant mutant R57 are highlighted in bold. The large and low frequency cohort a2 is omitted from the figure for clarity and brevity. See Additional file 3: Figure S1 for full dataset
Correlated evolutionary trajectories suggest genetic hitchhiking
All the 105 SNPs have their origin in either the MATa (33 SNPs) or MATα (72 SNPs) parental strains (Fig. 2 and Additional file 3: Figure S1). Visual examination of the allele frequencies suggested that SNPs could be further clustered in cohorts with strongly correlated evolutionary trajectories. For example, mutations aro1-CC1283-4TT and ste5-C512T, both on chromosome IV, display similar frequencies at all time points and originate in the MATa population. This observation suggested the existence of subgroups of SNPs of common origin hitchhiking on a few driver mutations. To test this hypothesis and identify cohorts of SNPs potentially linked by origin, we performed hierarchical clustering on the evolutionary trajectories of all SNPs (Fig. 2 and in Additional file 3: Figure S1). Nine cohorts of mutations were deduced from the resulting dendrograms. The majority of mutations are found at very low frequency (≤ 0.02) in all sampled time points, with varying levels of apparent selection. Those mutations are assigned to cohorts α1 and a2.
Three mutations show unique trajectories. Mutation mal11-G310A stands out as displaying the strongest apparent selection, with a mean allele frequency change of 1.69. Similarly, the gdh1-G47A mutation, with a mean allele frequency change of 1.53, is not clustered with other mutations. The gdh1-A68G mutation, displaying one of the strongest apparent selections (M = 1.60), is placed with the α3 cluster by the algorithm, but its trajectory is markedly different from other mutations in that cohort. It has a much lower initial frequency and displays stronger positive selection. We, therefore, also assigned gdh1-A68G to its own cohort (α4).
The most frequent MATa-derived cohorts are a5 and a6. In both cohorts, mutations start at a frequency of approximately 0.03 and increase steadily to reach maximum frequency after 3 cycles of mating and selection (R3) followed by a decline in subsequent cycles (R4 and R5). With similar trajectories but varying allele frequencies, it is not clear whether SNPs clustered in a5 and a6 belong to two independent cohorts or a single group of hitchhikers.
Cohort α3 consists of seven SNPs remarkable for their high and virtually identical initial frequency. SNPs from this group display a frequency of ~ 0.88 in the MATα population, indicating that they nearly swept the population at early selection steps. This group benefits from the founder effect, remaining highly represented until the end of the experiment. The general pattern followed by all but one mutation (ubp7-T2466A) suggests an absence of selection, or a slow decline in frequency (M = 0.903–0.970). The mutation in gene UBP7 diverges from the rest of the group, on average increasing in frequency. Large increases in frequency in the early mating and selection cycles (0.49–0.74 between R2 and R3) are followed by a decline, amounting to a modest measure of apparent selection for ubp7-T2466A (M = 1.070).
Five SNPs were detected in all reads of their original mutant population; srb8-C3787G and art5-G454T in the MATa and mtm1-A943T, avl9-C1806G and sro77-G(-160)T in the MATα. The frequency of these mutations oscillates around 0.50 in all 5 genome shuffled populations, indicating relative neutrality with respect to the selected phenotype (Additional file 2). These two observations suggest that they spontaneously arose in the parental clones before UV mutagenesis.
The signature of genetic hitchhiking observed in our data indicates that a large proportion of the SNPs identified by sequencing arose together in a few founding individuals. From the list of mutations detected by population sequencing, a restricted subset of mutations is thus expected to contribute to the phenotype of interest.
Certain genes are mutation hotspots
We observed several independent occurrences of distinct point mutations mapping to the same genes. A systematic survey revealed eight genes to which more than one mutation could be mapped (Additional file 4: Figure S2). Four strongly correlated mutations in genes STE5 and ARO1 were excluded because their correlated evolutionary trajectories suggested that they resulted from the same mutation event. Among the 25 remaining SNPs, 9 escape detection in at least one of the sampled time points (Tab “All_mutations” in Additional file 2). Most hotspot mutations displayed low frequency, with the exception of four SNPs, one of which mapped to MAL11 and the three others to GDH1. Mutations mapping to these two genes, as well as those mapping to NRG1, display a pattern of strongly positive apparent selection (Additional file 4: Figure S2). Remaining hotspots, mapping to genes VHR1, SSN2, YHR045W, UBP1 and COS111 display either negative or nearly neutral selection.
The most remarkable hotspot maps seven non-synonymous mutations to GDH1. A strong positive selection was observed for most of these mutations and three of them rapidly rose to prominence in the course of evolution (Fig. 3a). Three mutations cluster at the N-terminus of Gdh1p and four at the C-terminus (Fig. 3b). Multiple sequence alignment of Gdh1p homologs did not suggest a high level of sequence conservation for the affected residues (data not shown), but mapping of Gdh1p substitutions on a homology model (see Additional file 5) showed that both N- and C-terminal substitutions are grouped near the hinge region separating the two structural domains of the protein (Fig. 3c). Together, these observations convincingly argue for the strong selection of gdh1 alleles by our genome shuffling experiment.
Mutations in GDH1 played a critical role in the evolution of SSL tolerance. a Seven independent mutations result in amino acid substitutions in the Gdh1 protein. Their evolutionary trajectories cluster into a high (blue) and low (red) frequency group. b The high-frequency group maps at the N-terminus of Gdh1p, while the low frequency cohort affects residues clustered at the C-terminus. c Mapping of the seven substitutions onto a homology model of Gdh1p reveals that they all cluster in or near the hinge connecting the two structural domains of the protein, an area rich with known ubiquitination sites (displayed in orange). d Introduction of the gdh1 mutations into haploid and diploid wild-type backgrounds leads to a loss of fitness. Removal of mutant gdh1 alleles from the SSL-tolerant R57 background is also associated with a loss of tolerance. Error bars represent ± 1 standard deviation
To test the effect of gdh1 mutations on the phenotype, the seven-point mutants were reconstructed in a wild-type parental haploid background. Under non-selective conditions, with the exception of the crippling D411S mutation, all haploid gdh1 mutants grow as well as their wild-type parent but showed decreased fitness upon expose to the inhibitory substrate (Fig. 3d). Reversion of the S16F or F23S mutations found in strain R57 did not alter growth in permissive conditions, but resulted in loss of fitness in a wild-type homozygous background (Fig. 3d). In R57, the GDH1/GDH1 genotype is associated with a growth defect both in the presence and absence of inhibitors. Mutations in gene GDH1 are important for the inhibitor tolerance phenotype of R57, with reduced tolerance most obvious when S16F is reverted either alone or in combination with F23S. Together, these observations suggest reciprocal sign epistasis between gdh1 alleles and the wild-type and mutant backgrounds.
Effect of individual mutations on inhibitor tolerance phenotype
To estimate the contribution of individual mutations on the selected phenotype, we backcrossed random haploid segregants of R57 with the wild-type MATα parent strain and the fitness of 86 second-generation derivatives was assayed under permissive and inhibitory conditions. The result revealed a continuous distribution in their level of tolerance, with some mutants displaying a phenotype superior to the R57 parent (Additional file 6: Figure S3). A Kolmogorov–Smirnov test (α = 0.05) suggests a normal distribution for the fitness of the isolates, in agreement with the hypothesis of a polygenic quantitative trait.
We used amplicon sequencing to genotype the 86 segregants. For the vast majority of strains and loci, we achieved depth of coverage well above 30 with an average of 794, enabling confident genotyping (Additional file 1: Table S2). Multiple linear regression analysis was applied separately on the haploid and diploid data sets to predict the effect of each SNP on the phenotype (summarized in Fig. 4a, Additional file 7: Figure S4). Fewer variables and more data points mean that we have higher confidence in the haploid model than in the diploid model (see Additional file 5). Accordingly, there is better agreement between predictions of the haploid model and measured phenotypes. In both haploids and diploids, the strongest predictor for enhanced tolerance is the nrg1-G137T mutation. Examination of the genotype heatmap in Fig. 4a shows a clear clustering of nrg1 isolates at higher inhibitor tolerance levels. In haploids, the gsh1-T(-73)A mutation is also strongly associated with a high tolerance phenotype, although the model proposes negative epistasis between the nrg1 and gsh1 alleles. Mutations ssa1-C91A, tof2-C2141T and gdh1-C47T are also associated with modest increases in haploid tolerance, albeit the effect of the latter is proposed to be enhanced by interaction with gsh1-T(-73)A. Deleterious for haploids are mutations mal11-C310T, ubp7-T2466A and especially sgo1-C575A. Mutation mal11-C310T has virtually no effect (p value = 0.254 that linear coefficient is equal to 0), but is proposed to interact with nrg1-G137T to further increase tolerance.
Mutations in NRG1 and GSH1 are the main determinants of hydrolysate tolerance. Multiple linear regression (a) on the genotypes of segregants of an SSL-tolerant mutant identifies mutations nrg1-G137T and gsh1-T(-73)A as the strongest predictors of high tolerance. This prediction is confirmed in point mutants (b), with both mutations conferring increased tolerance to SSL. The nrg1-G137T mutation also increases tolerance to acetic acid and hydrogen peroxide in haploids. Bars in the graphs report area under the growth curve for the indicated mutants recorded in the presence (solid bars) and absence (striped bars) of inhibitors. Error bars represent ± 1 standard deviation
Reconstruction and reversion of SNPs
To confirm the contribution of individual mutations to the tolerance phenotype, a subgroup of 27 was reconstructed into the MATα parental strain. In agreement with our backcross experiment, gsh1-T(-73)A and nrg1-G137T both increased fitness of wild-type haploids (Fig. 4b). In contrast, the same SNPs in hetero- and homozygous wild-type diploids did not increase fitness (Additional file 8: Figure S5). Fitness of haploid and diploid nrg1 gsh1 double mutants is not higher than that of the fittest mutant, suggesting that the effect of these SNPs is non-cumulative. Further supporting a contribution of these SNPs to the phenotype, reversion to wild-type abolished the enhanced phenotype of haploid single mutants (Additional file 9: Figure S6).
To narrow the physicochemical stresses to which nrg1 and gsh1 mutations conferred resistance, we compared the fitness of haploid single mutants with wild type and R57 cells exposed to acetic acid or hydrogen peroxide (Fig. 4b). The nrg1-G137T mutation bestowed increased fitness in the presence of both compounds, enabling faster growth than wild type and R57. The gsh1-T(-73)A mutation does not seem to confer the same advantage. Among mutations identified by population sequencing, a subset of the most positively selected was also introduced in the MATα haploid, but an increase in tolerance was not observed for those strains.
Each SNP was also reverted to wild type in strain R57 (Additional file 10: Figure S7A). At most loci, reversion did not cause a detectable loss of fitness. The only exception was reversion of the gdh1-G47A mutation, which led to a pronounced decrease in fitness in the R57, R57 GDH1-68 and R57 SGO1 backgrounds. The loss of fitness associated with reversion of gdh1 alleles in the R57 background contrasts with the deleterious effect of these mutations in wild-type backgrounds (Fig. 3) and indicates reciprocal sign epistasis. We hypothesized that gdh1 alleles complement secondary deleterious mutations found in R57. We further reverted single mutations to wild type in the GDH1 or SGO1 gdh1-T68G backgrounds to identify those that would rescue the growth defects. The R57 GDH1 growth defect could not be rescued by reversion of single secondary SNPs. While we could not fully reproduce the R57 SGO1 gdh1-T68G growth defect, possibly because of batch-to-batch variations in hydrolysate composition, we identified two double revertants with reduced tolerance (Additional file 10: Figure S7B). Removal of mutation ynl058c-A7G in this background confers wild-type tolerance to SSL. Consistent with other observations, reversion of nrg1-G137T also leads to a loss of tolerance in the R57 SGO1 gdh1-T68G background. This effect is not observed in the R57 GDH1 background because loss of both gdh1 alleles leads to a loss of fitness that is too important to observe the effect of reverting to NRG1 or YNL058C. The partial loss of fitness in the homozygous gdh1-A68G derivative of R57 leaves room to observe the additional effect of reverting other mutations. Together, these results show that gdh1 alleles exert their effect via an epistatic mechanism. The precise mechanism is still elusive, but from the growth defect of R57 GDH1 cells in the presence and absence of inhibitors, we speculate that they act by complementation in a complex network of genetic interactions.
Discussion
The results of this study identified key molecular determinants of hydrolysate tolerance. Our evidence identifies genes NRG1, GSH1 and GDH1 as having the strongest impact on the selected phenotype. Below, we discuss their significance along with hypotheses on the contribution of other mutant alleles identified in this study. Knowledge on the genetic architecture of inhibitor-tolerant mutants also informs our understanding of the evolutionary determinants that dictated the outcomes of our genome shuffling experiment. Further below, we discuss these evolutionary determinants and how they may impact the outcomes of genome shuffling experiments.
Molecular determinants of fitness in lignocellulosic hydrolysates
Our evidence indicates that the low frequency but strongly selected mutation nrg1-G137T confers the strongest direct gains in hydrolysate inhibitor tolerance among all SNPs considered in this study (Fig. 4, Additional file 4: Figure S2, Additional file 7: Figure S4, Additional file 9: Figure S6 and Additional file 10: Figure S7). These results are consistent with previous RNAseq results in R57 that showed considerable upregulation of five targets of transcriptional regulator Nrg1p, including NRG1 itself [6].
NRG1 and its close paralog NRG2 encode DNA-binding proteins first identified as mediators of glucose repression [26, 27]. The Nrg1/2p repressors have been implicated in the response to various stresses, including glucose [28] and zinc limitation [29], alkaline pH [30], salt tolerance [31, 32] and organic acid challenge [33]. Identification of transcripts with altered expression in null mutants of NRG1 and NRG2 established their role in the regulation of the general stress response [34]. Deletion of NRG1 or NRG2 changes the transcription of 150 genes, many of which display stress response elements (STREs) or related sequences in their promoter regions. A significant overlap between the Nrg1/2p and Msn2/4p regulons further supports a role for Nrg proteins in the regulation of the general stress response [26, 35]. We, therefore, propose that nrg1-C137A is a loss of function mutation that leads to the upregulation of general stress response genes.
Our models predict that mutation gsh1-T(-73)A makes the second largest contribution to inhibitor tolerance in haploids (Fig. 4a). Accordingly, haploids carrying this single mutation display enhanced growth in the presence of high concentrations of hydrolysate (Fig. 4b). The near sweep of the MATα mutant pool by the gsh1-T(-73)A allele also suggests a significant selective advantage in the presence of high concentrations of hydrolysate. We have previously shown that haploids carrying the mutant gsh1-T(-73)A allele accumulate lower levels of reactive oxygen species (ROS) than their wild-type parents when exposed to high concentrations of hydrolysate [36]. This is consistent with the role of Gsh1p in the synthesis of the antioxidant glutathione [37]. The mutation identified in our mutants is located 73 bp upstream of the start codon, outside Yap1p and other hydrogen peroxide responsive sequences [38,39,40]. The position of this SNP in the region proximal to the start codon identifies alteration of the basal promoter as the most likely mechanism. We propose that upregulation of GSH1 by this modified promoter increases glutathione synthesis and reduces accumulation of ROS in gsh1-T(-73)A mutants.
Several lines of evidence point to GDH1 as a key determinant of selection in our genome shuffling experiment. Notably, this gene is the most populated mutation hotspot (Fig. 3a) and gdh1 alleles appear required for hydrolysate tolerance in mutant R57 (Fig. 3d). However, their effect appears epistatic, since their introduction into wild-type backgrounds is associated with growth defects, perhaps suggesting a compensatory role in hydrolysate tolerant strain R57 (Fig. 3d).Glutamate dehydrogenase Gdh1p, along with close homolog Gdh3p, catalyzes amination of α-ketoglutarate, yielding glutamate [41]. Under fermentative conditions, Gdh1p is the dominant glutamate dehydrogenase, while carbon limitation, non-fermentable carbon sources and entry into the stationary phase induce the expression of Gdh3p [42, 43]. Transcription of GDH1 is sustained at all phases of growth, but entry into the stationary phase triggers ubiquitin-mediated degradation of Gdh1p. Gdh3p is specifically expressed during the stationary phase. Gdh1p is a faster enzyme, suited for growth-sustaining glutamate synthesis. Gdh3p is slower, better suited to sustain glutathione synthesis during the stationary phase and under stressful conditions [42]. Accordingly, Gdh3p has been implicated in stress tolerance in yeast, while transient loss of tolerance to hydrogen peroxide is observed during early phases of growth in gdh1Δ mutants [43]. From the mapping of the seven amino acid substitutions on the Gdh1 protein, we suggest two potential mechanisms for their action. One possibility is that they affect inter-domain flexibility and, thus, catalytic activity, as suggested by structural studies of bacterial homologs [44]. The substitutions are located near known ubiquitination sites [43, 45]. A second hypothesis is thus that they impair degradation of Gdh1p in conditions of stress or during the stationary phase. Regardless of its underlying mechanism, we propose that the gdh1 hotspot was selected to compensate the pull on glutamate exerted by upregulation of glutathione biosynthesis in gsh1-T(-73)A mutants, especially under hydrolysate inhibitor stress. This hypothesis would explain the strong growth defect incurred by R57 upon reversion of gdh1 mutations and, therefore, the reciprocal sign epistasis observed with these mutations.
The role of MAL11 in the transport of the stress-protectant molecule trehalose suggests involvement of mutations of this hotspot in hydrolysate tolerance [46]. Our model of hydrolysate tolerance in haploids suggests interactions between the nrg1-C137A and mal11-C310T mutations. MAL11 is repressed by glucose, notably via the action of Mig1p [47,48,49]. Overlap between the Mig1p and Nrg1p regulons suggests a potential mechanism for this regulation [26].
Reversion of the ynl058c-A7G mutation in R57 does not cause a detectable loss of tolerance to the hydrolysate inhibitors. However, in a derivative of R57 wild type at the SGO1 locus and homozygous for the gdh1-A68G allele, reversion to wild type at the YNL058C locus leads to a loss of SSL tolerance. This suggests a role for this gene in inhibitor tolerance. The function of YNL058C is essentially unknown, but the protein it encodes appears to localize to the vacuole [50]. Both YNL058C and its paralog PRM5 are induced via the cell wall integrity pathway, indicating a role in the response to cell wall damage [51, 52]. Downregulation of YNL058C was also observed upon DNA damage [53]. Together, these reports indicate a role for YNL058C in the response to stress and cell damage, in agreement with an involvement in hydrolysate tolerance.
Mutation ste5-C512T displays the highest apparent selection and average frequency in cohort a6. The model of R57 backcrosses indicates that it makes the second highest contribution to inhibitor tolerance in diploids (Additional file 7: Figure S4). STE5 encodes a scaffold protein involved in facilitation and integration of pheromone-induced MAPK signaling [54]. Components of the pheromone-induced MAPK pathway were also implicated in stress signaling [55,56,57,58,59]. Because Ste5p binds the Kss1p MAPK, it mediates signals involved in stress. It is thus likely that the ste5-C512T mutation modulates the Ste20p-Ste11p-Ste7p-Kss1p pathway to stimulate the execution of a stress response program.
Our understanding of the hydrolysate inhibitor tolerance phenotype selected in our genome shuffling mutants, as described above, is summarized in Fig. 5. In brief, our results indicate that mutant alleles nrg1-G137T and gsh1-T(-73)A are the main determinants of hydrolysate inhibitor tolerance in our evolved mutants. Our evidence also ascribes a critical role for gdh1 alleles. The phenotype associated with these alleles in parental and mutant strains demonstrates reciprocal sign epistasis and is coherent with a compensatory mechanism. We propose putative mechanisms for other potential contributing alleles. These alleles may help in the rational engineering of hydrolysate inhibitor tolerance strains. However, a recent study showed that the genetic determinants of hydrolysate inhibitor tolerance in yeast can prove highly strain-dependent [60]. Therefore, it would be interesting to test the impact of mutations identified herein in various industrial strain backgrounds.
Network map model of SSL tolerance in genome shuffled mutants. Nodes represent mutations or phenotypes and edges represent effects or interactions. Curvature of the edges is clockwise with respect to effector nodes and counter-clockwise with respect to target nodes. Green edges indicate a stimulating or enhancing effect, while red lines indicate an inhibition of the target. Orange lines indicate persisting doubts on the relationship between the nodes that they connect. Thickness of the edges is related to the amount of evidence available for the indicated interactions
Evolutionary determinants of selection by genome shuffling
Our experiment is characterized by a set of early genetic features that had a determining effect on its overall outcomes. These features are the presence of mutations in the parental strains, prevalent genetic hitchhiking, and a selective sweep that severely reduced diversity at early selection steps. We argue that these features favored specific evolutionary solutions and elicited a strong selective pressure in favor of compensatory mechanisms.
The MATα parental strain contained mutations mtm1-A943T, avl9-C1806G and sro77-G(-160)T, while the MATa parent carried SNPs srb8-C3787G and art5-G454T. We draw parallels between these founding mutations and mutational hotspots identified by population genome sequencing. Both MTM1 and YHR045W have been implicated in iron metabolism [61,62,63]. Similarly, the ssn2 hotspot can be linked to the srb8-C3787G substitution. Both genes encode subunits of the RNA polymerase II mediator complex [64,65,66,67,68]. The ubp1 hotspot is notable, owing to the high frequency of the ubp7-T2466A mutation in our pool. Both genes encode ubiquitin-specific proteases [69, 70]. The prevalence of the founding art5-G454T mutation, mapping to a gene involved in the regulation of membrane protein homeostasis [71], suggests that complementing mutations involved in this cell process were selected during our experiment. The yhr045w, ssn2 and ubp1 mutations all remain at low frequency, with either weakly positive or negative apparent selection. This would suggest that complementation of founder mutations confers marginal competitive advantages. However, the prevalence and persistence of mutation ubp7-T2466A are hypothesized to result from the same epistatic dynamics. The coincidental presence of this mutation in the same mutant as the α3 driver gsh1-T(-73)A likely caused the selection of ubp7-T2466A by hitchhiking, while its arguably minor role in complementing art5-G454T was seemingly sufficient to ensure its persistence, in contrast with other hitchhikers of the same cohort.
Clustering of mutations in cohorts with correlated evolutionary trajectories is commonly observed in experimental evolution experiments as a signature of genetic hitchhiking [14]. In the context of our genome shuffling experiment, this observation suggests that a large proportion of mutant alleles arose together in a few founding individuals. Prominent examples are cohorts a5, a6 and α3, putatively driven by ste5-C512T or gdh1-G299A, and gsh1-T(-73)A, respectively (Fig. 2). We expect that most examples of genetic hitchhiking negatively affected the fitness of mutants, because the majority of mutations tend to be neutral or deleterious [72]. Previous studies in yeast on the evolutionary effects of sexual recombination suggest that it favors purifying selection of hitchhikers [16]. Accordingly, the majority of putative hitchhikers in our experiment either display consistently low frequency or negative apparent selection (Fig. 2). Despite evidence of purifying selection, putative hitchhikers of cohort α3 persist at high frequency until the end of the GS experiment. This could in part be due to vegetative cells escaping the sporulation and mating process. Yet, sporulation efficiency was high (40% on average) and both digestion and sonication of vegetative cells after sporulation should have minimized the impact of non-shuffled mutants on the evolutionary dynamics of the experiment. Further, cohort α3 nearly swept the initial pool of MATα mutants likely due to an aggressive early selection. This first selection was performed before the onset of sexual recombination cycles and illustrates the well-documented effect of clonal interference on the evolution of asexually reproducing populations [14, 16].
We propose that the founding features discussed above caused major evolutionary responses, and that those are critical to understanding the dynamics of the experiment. For example, the presence of mutational hotspots identifies loci under strong selective pressure and we have shown that many appear related to founding mutations. Similarly, the mechanism proposed above for epistasis between gdh1 and gsh1 mutant alleles would explain the evolutionary signal detected at the GDH1 hotspot. The convergence of the gsh1-driven selective sweep with founding mutation art5-G454T also appears to have driven the persistence of the ubp7-T2466A hitchhiker allele. Finally, the minor evolutionary role played by the tolerance-conferring nrg1-G137T mutation may be caused at least in part by the founder effect favoring gsh1-T(-73)A coupled to negative epistasis between the two alleles, as suggested by our linear model of hydrolysate tolerance (Fig. 4a). This observation resonates with experimental evolution studies of sexually reproducing yeast that showed a major role for standing diversity and ascribed a minor evolutionary role to rare or de novo mutations [23]. It also illustrates the dominant role played by compensatory mechanisms over direct enhancement of hydrolysate tolerance in our experiment.
In contrast to previous experimental studies of compensatory evolution, the examples proposed in this study rely on convergent signals at specific loci. Convergent compensatory evolution at the functional rather molecular level has previously been reported [73]. Because the path of compensatory evolution was shown to be constrained both by environmental factors and the genetic background, the level of molecular convergence that we observe would indicate a targeted response to highly specific conditions [74].
Descriptive model of evolutionary dynamics in the genome shuffling experiment
Based on the proposed model of SSL tolerance and on the allele frequency time series obtained from population sequencing (Fig. 2), we propose a model to describe the evolutionary dynamics of our genome shuffling experiment. Mutagenesis generated equally diverse pools of mutants from the MATα and MATa parental strains. An aggressive selection regimen restricted genetic diversity in the MATα pool, leading to a near sweep by mutants carrying the gsh1-T(-73)A allele and α3 hitchhikers. Mutations mal11-A482T and gdh1-A68G were found at a low frequency in this pool. A more relaxed selection generated a more diverse pool of MATa mutants, among which were tolerance enhancing nrg1-C137A and ste5-C512T mutations. Cohorts a6 and perhaps a5 hitchhiked on this latter mutation. The gdh1 mutations (with the exception of A68G) and mal11-G310A were also selected into this initial pool. Initial recombination created the first epistatic pairs between tolerance-conferring and compensatory mutations. Combinations of founders with their complementing mutations also occurred on a large scale at this stage. Selection on these shuffled mutants brought a large increase in the frequency of complementing mutations; thanks to the competitive advantage they imparted onto SSL-tolerant but metabolically imbalanced mutants. Further shuffling and selection brought several of these epistatic relationships into single cells, increasing their fitness in the presence of SSL. The strong selective advantage of nrg1-C137A and perhaps its separation from deleterious alleles to which it was initially linked led to its steady increase in frequency from a low initial frequency. Additional rounds of shuffling could have witnessed the rise to prominence of this nrg1 allele.
Conclusions
In this study, we identified some of the key determinants of selection in a genome shuffling experiment. We ascribed critical roles to loci NRG1, GSH1 and GDH1 to the hydrolysate inhibitor tolerance phenotype. With regard to recent results [60], the applicability of these evolutionary solutions to different strain backgrounds remains to be established.
More fundamentally, we have illustrated the impact that a few, early features can have on the course of evolutionary engineering by genome shuffling. It also suggests that this regimen of recursive recombination leads to the widespread selection of compensatory mechanisms, illustrating the construction of strains in which delicate complementation networks operate to offset the fitness cost incurred by founding mutations, hitchhikers and the pleiotropic effects of core beneficial alleles. To expand on the specific outcomes of our experiment, we propose that purposefully designed genome shuffling experiments performed on diverse genetic backgrounds with precise variations in initial conditions could aid the design of future strain development endeavors, and contribute to our understanding of evolution in sexually reproducing populations, especially as it pertains to compensatory evolution.
Methods
Evolution by genome shuffling
All strains of S. cerevisiae used in this study are derived from prototrophic strains CEN.PK113-1A (wild-type MATα) and CEN.PK113-7D (wild-type MATa). Mating of the haploids was used to generate the wild-type diploid. The genome shuffling experiment is described in detail in a previous publication [24] and is summarized in Fig. 1. Briefly, to generate genetically diverse starting populations, MATα and MATa haploid mutant pools were generated by UV irradiation. These initial pools were spread onto gradient plates for selection of cells with inhibitor tolerance above the wild type. Irradiated cells growing above the wild type were scraped off the plate and aliquots from both populations were preserved as glycerol stocks at − 80 °C for later sequencing. The two populations of selected haploid were mixed 1:1 for mating and diploids were selected on gradient plates. This first population of selected diploids was sporulated, digesting vegetative cells with Zymolyase (MP Biomedicals) and sonicating them before mating, generating a first genome shuffled population R1. Four additional recursive cycles of selection, sporulation and mating were performed resulting in populations R2–R5, which showed increasing tolerance to the inhibitory substrate at each mating cycle. Genome shuffled populations selected on gradient plates were propagated overnight in YPD at 30 °C and aliquots were preserved as glycerol stocks at − 80 °C for later sequencing.
Pool-seq of evolved populations
Prior to genomic DNA extractions, cells from glycerol stocks of the two UV irradiated (a-UV and α-UV) and five genome shuffled populations (R1–R5) were thawed and incubated in YPD for 1 h. They were then suspended in 50 mM Tris–HCl pH 8.0,10 mM EDTA, 5% 2-mercaptoethanol (v/v), 200 U/ml yeast lytic enzyme (MP Biomedicals) for 1 h at 37 °C. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions and quantified using the QuantiFluor dsDNA System (Promega). Genomic DNA was sequenced at the McGill University and Genome Quebec Innovation Centre using the TrueSeq library preparation reagents and an Illumina HiSeq 2500 sequencer (100 bp paired-end reads). Each of the 7 populations was sequenced on a separate lane of a HiSeq chip to maximize depth of coverage.
Quality control of raw sequencing data was performed using FastQC [75] and overlapping read pairs were merged with PEAR [76]. Alignment to the CEN.PK113-7D reference genome [25] was done using bwa mem [77] and performed separately for overlapping and non-overlapping reads. Output SAM files for overlapping and non-overlapping reads were merged with the MergeSamFiles utility in Picard Tools [78]. Picard was next used to add read groups, sort reads, then mark and remove duplicates prior to indel realignment with the Genome Analysis Toolkit [79,80,81]. Alignment metrics were extracted using Picard Tools.
SNPs were called and filtered using a base error model inspired from Barrick and Lenski [18] as detailed in Additional file 5.
Mutant allele frequency, strength of selection and hierarchical clustering of evolutionary trajectories
The proportion of mismatch reads at given genomic coordinates was considered to reflect the frequency (p) of the associated mutant allele within the sequenced populations. The frequency of all mutations was extracted for the seven sequenced populations. The frequency of each mutation was necessarily zero in one of the haploid populations, allowing the identification of its origin (MATα or MATa). Furthermore, because the two haploid populations represented a single evolutionary time point, their frequency was averaged to obtain the pre-shuffling allele frequencies (i.e., pUV = (pα + pa)/2). Allele frequencies of the R1–R5 populations each represented time points of their own.
Strength of the positive or negative selection was estimated from allele frequency change across the time points (Δpt1−t2 = pt2/pt1). The geometric mean frequency change (noted M) was used as a synthetic measure of selection to smooth the effect of proportionally large frequency changes often observed between time points UV and R1. Hierarchical clustering of evolutionary trajectories was performed by running the clustermap routine of the Seaborn Python library [82].
Backcrossing experiment
To assess the contribution of each SNP to the inhibitor tolerance phenotype, strain R57 was backcrossed with parental wild-type MATα. Cells from the resulting F2 population were scored for tolerance to inhibitors (Additional file 6: Figure S3). Haploids of R57 were generated on sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% dextrose, 2% agar) incubated at room temperature for 6 days, resulting in > 50% sporulation efficiency. Cells were scraped from the plate and digested overnight at 30 °C with 100 U of yeast lytic enzyme (MP Biomedicals) in 5 ml of H2O containing 10 μl of 2-mercaptoethanol. Five microlitre of 1.5% IGEPAL was added to the digest, incubated on ice for 15 min and sonicated 3× for 30 s. Spores were harvested by centrifugation at 12,000×g for 10 min and suspended in 5 ml 1.5% IGEPAL. The sonication procedure was repeated once more and the spores were suspended in 250 μl of YPD broth. For mating, spores were mixed with an approximately equal amount of MATα cells (CENPK113-1A), spotted on YPD agar and incubated overnight at 30 °C. Resulting R57 × 1A spores were spotted on YPD agar and then allowed to germinate and mate for 48 h. The resulting diploids were submitted to a second round of sporulation and mating to further shuffle mutations. After mating, cells were streaked for single colonies on YPD agar and 86 isolated colonies were picked for genotyping and fitness assays.
Preparation of yeast genomic DNA template for PCR
Cells from 1.5 ml of culture were harvested in a tabletop microcentrifuge and the resulting pellet was suspended in 250 μl of 50 mM Tris–Cl pH 8.0 supplemented by 20 U of yeast lytic enzyme. This digestion solution was incubated at 37 °C for 1 h. Cell lysis was induced by adding 250 μl of 200 mM NaOH, 1% SDS, vortexing, and incubating for 5 min at room temperature. The lysates were neutralized with the addition of 350 μl of 3 M potassium acetate pH 5.5 and clarified by centrifugation at 13,300×g for 10 min. DNA from the resulting supernatants was precipitated by adding 600 μl of 2-propanol and centrifuged at 13,300×g for 10 min. The resulting DNA pellet was air dried for 15 min, suspended in 100 μl of H2O, and vortexed before incubation for 15 min in a water bath at 55 °C.
Genotyping of backcrossed isolates
We genotyped 86 backcrossed isolates at 18 of the mutant loci identified in R57 by PCR amplicon sequencing. Both forward and reverse primers consisted of a common 5′ heel sequence (forward: 5′-CGTTCAACCTTGTCCAACAGTG-3′ and reverse: 5′-GAAGCGATGACTCGAGCGTATT-3′) and a 24–28 nucleotide gene-specific sequence at the 3′ end. PCRs contained genomic DNA template, 0.5 μM primers, 200 μM dNTPs, 1× high fidelity Phusion buffer (Thermo Fisher), 1.5% DMSO and 1 U of Phusion High Fidelity DNA polymerase (Thermo Fisher) in 50 μl. Cycling conditions were as follows: 98 °C for 30 s, then 35 cycles of 98 °C for 10 s, 55 °C for 20 s, 72 °C for 4 s, followed by a 5 min final extension at 72 °C. Ion torrent sequencing adapters and barcodes were added in a second PCR. This second reaction was performed using a common reverse primer consisting of the ion torrent P1 adapter (5′-CCTCTCTATGGGCAGTCGGTGAT-3′) and the reverse heel sequence. The 96 forward primers consisted of the ion torrent A adapter (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′), a unique 8 nucleotide barcode and the forward heel sequence. Composition of this second PCR was the same as for locus-specific amplification, using 1 μl of a 1:10 dilution of the first reaction as template. Cycling conditions were: 98 °C for 30 s, then 35 cycles of 98 °C for 10 s, 70 °C for 20 s, 72 °C for 6 s, followed by a 5 min final extension at 72 °C. All primers used for genotyping are listed in Additional file 1: Table S3. PCR products carrying the same barcode were pooled and DNA migrating between 100 and 400 bp was purified on a 2% agarose gel using the GeneJet gel extraction kit (Thermo Fisher). Equal amounts of all pools were mixed to make the sequencing library. The DNA concentration of this pool was measured on a Qubit 2.0 fluorometer (Life Technologies). Ion torrent sequencing template was prepared from our pooled library with an Ion PGM Template OT2 400 Kit according to the manufacturer’s instructions. Sequencing was performed with an Ion PGM sequencer using the Ion PGM Hi-Q sequencing kit and an Ion 316 Chip v2 BC, all following manufacturer’s protocols and instructions.
Reads from the Ion PGM were sorted and trimmed using a custom python script, and then aligned to the CEN.PK113-7D reference genome [25] using bwa mem [77]. Read counts for each of the SNP positions were extracted with bam-readcount [83] setting both minimum mapping quality and minimum base quality at 30. Genotypes were called from read counts as follows: heterozygotes were distinguished from homozygotes by assuming that in homozygotes, the most frequent base call would have a frequency of 0.997 and all other base calls 0.001 each. A G test was performed to test whether the base count distribution differed significantly from this assumption. If it did, a heterozygous genotype was called. Otherwise, the identity of the most frequent base call was checked. If the most frequent base call was the reference base, a wild-type genotype was called, otherwise a homozygous mutant genotype was called. In strains previously identified as haploid, heterozygosity calls were corrected and genotype was called based on the identity of the most frequent base call. All genotype calls were reviewed visually.
Reconstruction and reversion of SNPs using CRISPR-Cas9
To probe their contribution on the observed phenotype, point mutations were reintroduced in wild-type backgrounds (CEN.PK113-1A, MATα or CEN.PK113-7D, MATa) or reverted to wild type in strain R57 using CRISPR-Cas9 as described previously [36]. Briefly, gRNAs were designed to introduce double-stranded breaks (DSBs) in the vicinity of the mutation. DSBs were repaired by homologous recombination with donor DNAs replacing the 20-nt protospacer sequence by a heterologous stuffer sequence. This stuffer was targeted by a second gRNA and the resulting DSBs were repaired by a stuffer-free donor DNA carrying the point mutations. The result was the seamless introduction of single nucleotide changes.
Fitness assays
Cell growth when exposed to varying concentrations of the inhibitory substrate was used to measure the fitness of the strains. Prior to the assays, cells we pre-adapted with overnight incubations in undiluted inhibitory substrate at 30 °C. For fitness assays, adapted cells were washed 3× in 10 mM sodium citrate pH 5.5 and inoculated to a final concentration of ~ 2 × 105 into YNB 1% glucose supplemented with varying concentrations (0–85%) of the inhibitory substrate in 96-well plates (Costar 3595, Corning). Plates were incubated at 30 °C with shaking in a Tecan Sunrise absorbance reader, measuring absorbance at 595 nm every 20 min. Because inhibitors may affect growth rate, lag time, maximum cell density or any combination thereof, we used the area under the growth curves as a measure of fitness. The lignocellulosic inhibitory substrate was kindly supplied by Tembec (Témiscaming, Québec) and AV Cell (Atholville, New Brunswick). The pH of the substrate was adjusted to 5.5 with 10 M NaOH prior to use in culture medium.
Multiple linear regression model
Detailed description of multiple linear regression methodology is provided in Additional file 5. Briefly, contribution of individual SNPs to the phenotype was predicted from linear models. These models were built by multiple linear regression, using genotype at each of the mutant loci as explanatory variables and area under growth curves in 85% inhibitor substrate as the response variable. Distinct models were built for haploid and diploid mutants. A single binary variable was used for each locus in haploids, with wild type = 0 and mutant = 1. In diploids, each locus was represented by three binary variables representing the wild type, heterozygous mutant and homozygous mutant genotypes, respectively. For the haploid model, fitting using all possible combinations of variables was performed and the model that minimized Mallows Cp and variance of residuals was chosen. The large number of variables involved with diploids made this approach impractical and so it was performed in a stepwise manner. Interaction between loci was modeled in an ad hoc manner, using a stepwise methodology similar to what was used for diploids.
References
Biot-Pelletier D, Martin VJJ. Evolutionary engineering by genome shuffling. Appl Microbiol Biotechnol. 2014;98:3877–87.
Wei Y, Wang C, Wang M, Cao X, Houa L. Comparative analysis of salt-tolerant gene HOG1 in a Zygosaccharomyces rouxii mutant strain and its parent strain. J Sci Food Agric. 2013;93:2765–70.
Yin H, Ma Y, Deng Y, Xu Z, Liu J, Zhao J, et al. Genome shuffling of Saccharomyces cerevisiae for enhanced glutathione yield and relative gene expression analysis using fluorescent quantitation reverse transcription polymerase chain reaction. J Microbiol Methods. 2016;127:188–92.
Zhao J, Zhang C, Lu J, Lu Z. Enhancement of fengycin production in Bacillus amyloliquefaciens by genome shuffling and relative gene expression analysis using RT-PCR. Can J Microbiol. 2016;62:431–6.
Zheng D-Q, Chen J, Zhang K, Gao K-H, Li O, Wang P-M, et al. Genomic structural variations contribute to trait improvement during whole-genome shuffling of yeast. Appl Microbiol Biotechnol. 2014;98:3059–70.
Pinel D, Colatriano D, Jiang H, Lee H, Martin VJ. Deconstructing the genetic basis of spent sulphite liquor tolerance using deep sequencing of genome-shuffled yeast. Biotechnol Biofuels. 2015;8:53.
Harner NK, Bajwa PK, Formusa PA, Austin GD, Habash MB, Trevors JT, et al. Determinants of tolerance to inhibitors in hardwood spent sulfite liquor in genome shuffled Pachysolen tannophilus strains. Antonie Van Leeuwenhoek. 2015;108:811–34.
Huang S, Xue T, Wang Z, Ma Y, He X, Hong J, et al. Furfural-tolerant Zymomonas mobilis derived from error-prone PCR-based whole genome shuffling and their tolerant mechanism. Appl Microbiol Biotechnol. 2018;102:3337–47.
Zhang Y, Liu J-Z, Huang J-S, Mao Z-W. Genome shuffling of Propionibacterium shermanii for improving vitamin B12 production and comparative proteome analysis. J Biotechnol. 2010;148:139–43.
Zhao J, Cao L, Zhang C, Zhong L, Lu J, Lu Z. Differential proteomics analysis of Bacillus amyloliquefaciens and its genome-shuffled mutant for improving surfactin production. Int J Mol Sci. 2014;15:19847–69.
Guan N, Shin H, Chen RR, Li J, Liu L, Du G, et al. Understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the level of proteomics. Sci Rep. 2014;4:6951.
Long A, Liti G, Luptak A, Tenaillon O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat Rev Genet. 2015;16:567–82.
Jerison ER, Desai MM. Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Curr Opin Genet Dev. 2015;35:33–9.
Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–4.
Kvitek DJ, Sherlock G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 2011;7(4):e1002056.
Mcdonald MJ, Daniel P, Rice DP, Desai MM, Daniel P. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. 2016;531:233–6.
Voordeckers K, Kominek J, Das A, Espinosa-Cantú A, De Maeyer D, Arslan A, et al. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 2015;11(11):e1005635.
Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009;74:119–29.
Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–70.
Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–61.
Herron MD, Doebeli M. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 2013;11(2):e1001490.
Cooper VS, Staples RK, Traverse CC, Ellis CN. Parallel evolution of small colony variants in Burkholderia cenocepacia biofilms. Genomics. 2014;104:447–52.
Burke MK, Liti G, Long AD. Standing genetic variation drives repeatable experimental evolution in outcrossing populations of Saccharomyces cerevisiae. Mol Biol Evol. 2014;31:3228–39.
Pinel D, D’Aoust F, del Cardayre SB, Bajwa PK, Lee H, Martin VJ. Saccharomyces cerevisiae genome shuffling through recursive population mating leads to improved tolerance to spent sulfite liquor. Appl Environ Microbiol. 2011;77:4736–43.
Otero JM, Vongsangnak W, Asadollahi MA, Olivares-Hernandes R, Maury J, Farinelli L, et al. Whole genome sequencing of Saccharomyces cerevisiae: from genotype to phenotype for improved metabolic engineering applications. BMC Genomics. 2010;11:723.
Park SH, Koh SS, Chun JH, Hwang HJ, Kang HS. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2044–50.
Zhou H, Winston F. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2001;2:5.
Kuchin S, Vyas VK, Carlson M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol. 2002;22:3994–4000.
Lyons T, Gasch A, Gaither L, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA. 2000;97:7957–62.
Lamb TM, Xu W, Diamond A, Mitchell AP. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem. 2001;276:1850–6.
Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:677–86.
Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–91.
Mira NP, Becker JD, Sá-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS. 2010;14:587–601.
Vyas VK, Berkey CD, Miyao T, Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1882–91.
Murad AMA, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–52.
Biot-Pelletier D, Martin VJ. Seamless site-directed mutagenesis of the Saccharomyces cerevisiae genome using CRISPR-Cas9. J Biol Eng. 2016;10:6.
Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol. 2001;39:533–41.
Stephen DW, Jamieson DJ. Amino acid-dependent regulation of the Saccharomyces cerevisiae GSH1 gene by hydrogen peroxide. Mol Microbiol. 1997;23:203–10.
Sugiyama KI, Izawa S, Inoue Y. The Yap1p-dependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae. J Biol Chem. 2000;275:15535–40.
Dormer UH, Westwater J, Stephen DWS, Jamieson DJ. Oxidant regulation of the Saccharomyces cerevisiae GSH1 gene. Biochim Biophys Acta Gene Struct Expr. 2002;1576:23–9.
Avendano A, Deluna A, Olivera H, Valenzuela L, Gonzalez A. GDH3 encodes a glutamate dehydrogenase isozyme, a previously unrecognized route for glutamate biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:5594–7.
DeLuna A, Avendaño A, Riego L, González A. NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae: purification, kinetic properties, and physiological roles. J Biol Chem. 2001;276:43775–83.
Lee YJ, Kim KJ, Kang HY, Kim HR, Maeng PJ. Involvement of GDH3-encoded NADP+-dependent glutamate dehydrogenase in yeast cell resistance to stress-induced apoptosis in stationary phase cells. J Biol Chem. 2012;287:44221–33.
Stillman TJ, Baker PJ, Britton KL, Rice DW. Conformational flexibility in glutamate dehydrogenase: role of water in substrate recognition and catalysis. J Mol Biol. 1993;234:1131–9.
Starita LM, Lo RS, Eng JK, von Haller PD, Fields S. Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics. 2012;12:236–40.
Han EK, Cotty F, Sottas C, Jiang H, Michels CA. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol Microbiol. 1995;17:1093–107.
Hu Z, Nehlin JO, Ronne H, Michels CA. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr Genet. 1995;28:258–66.
Zhang CY, Bai XW, Lin X, Liu XE, Xiao DG. Effects of SNF1 on maltose metabolism and leavening ability of baker’s yeast in lean dough. J Food Sci. 2015;80:M2879–85.
Hu Z, Yue Y, Jiang H, Zhang B, Sherwood PW, Michels CA. Analysis of the mechanism by which glucose inhibits maltose induction of MAL gene expression in Saccharomyces. Genetics. 2000;154:121–32.
Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91.
Arias P, Díez-Muñiz S, García R, Nombela C, Rodríguez-Peña JM, Arroyo J. Genome-wide survey of yeast mutations leading to activation of the yeast cell integrity MAPK pathway: novel insights into diverse MAPK outcomes. BMC Genomics. 2011;12:390.
Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol Microbiol. 1999;34:1049–57.
Maas N, Miller K, DeFazio L, Toczyski D. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–19.
Elion EA. The Ste5p scaffold. J Cell Sci. 2001;114:3967–78.
Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–300.
O’Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–86.
Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–4.
Roberts RL, Fink GR. Elements of a single map kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–85.
Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci USA. 1999;96:12679–84.
Sardi M, Paithane V, Place M, Robinson DE, Hose J, Wohlbach DJ, et al. Genome-wide association across Saccharomyces cerevisiae strains reveals substantial variation in underlying gene requirements for toxin tolerance. PLoS Genet. 2018;14(2):e1007217.
Luk E, Carroll M, Baker M, Culotta VC. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc Natl Acad Sci USA. 2003;100:10353–7.
Whittaker MM, Penmatsa A, Whittaker JW. The Mtm1p carrier and pyridoxal 5′-phosphate cofactor trafficking in yeast mitochondria. Arch Biochem Biophys. 2015;568:64–70.
Jo WJ, Kim JH, Oh E, Jaramillo D, Holman P, Loguinov AV, et al. Novel insights into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae. BMC Genomics. 2009;10:130.
Carlson M, Osmond BC, Neigeborn L, Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984;107:19–32.
Balciunas D, Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23:4426–33.
Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30(5):235–9.
Larschan E, Winston F. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol Cell Biol. 2005;25:114–23.
Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–20.
Tobias JW, Varshavsky A. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12021–8.
Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–92.
Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–25.
Ohta T. Near-neutrality in evolution of genes and gene regulation. Proc Natl Acad Sci USA. 2002;99:16134–7.
Szamecz B, Boross G, Kalapis D, Kovacs K, Fekete G, Farkas Z, et al. The genomic landscape of compensatory evolution. PLoS Biol. 2014;12(8):e1001935.
Filteau M, Hamel V, Pouliot M, Gagnon-Arsenault I, Dubé A, Landry C. Evolutionary rescue by compensatory mutations is constrained by genomic and environmental backgrounds. Mol Syst Biol. 2015;11(10):832.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 11 Oct 2018.
Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–20.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60.
Broad Institute. Picard: a set of command line tools for manipulating high-throughput sequencing (HTS) data and formats. 2016. http://broadinstitute.github.io/picard/. Accessed 11 Oct 2018.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1-33.
Waskom M. Seaborn: statistical data visualization. 2016. https://seaborn.pydata.org/. Accessed 11 Oct 2018.
Larson D. Bam-readcount: program to generate metrics at single nucleotide positions from BAM files. 2016. https://github.com/genome/bam-readcount. Accessed 11 Oct 2018.
Authors’ contributions
According to the CRediT taxonomy, author contributions are the following. Conceptualization: DBP, DP, KL, VM. Methodology: DBP, DP, KL, VM. Software: DBP. Validation: DBP. Formal analysis: DBP. Investigation: DBP, DP, KL. Resources: VM. Data curation: DBP. Writing—original draft preparation: DBP, VM. Writing—review and editing: DBP, DP, KL, VM. Visualization: DBP. Supervision: VM. Project administration: VM. Funding acquisition: VM. All authors read and approved the final manuscript.
Acknowledgements
We give our most sincere thanks to Hung Lee (University of Guelph) and Kevin Shiell (Collège communautaire du Nouveau-Brunswick) for helping us secure a supply of spent sulfite liquor. We thank Tembec and AV Cell for producing and graciously providing spent sulfite liquor for use in our research. We would also like to thank Mathieu Bourgey of the Canadian Centre for Computational Genomics for advice in the analysis of NGS data. We also thank Vladimir Rheinharz of the McGill University Department of Computer Science for advice on the critical assessment of multiple linear regression results.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study, with the exception of population sequencing data, are included in this published article and its supplementary information files. The datasets generated and analyzed during the current study are available in the National Center for Biotechnology Information (NCBI) sequence read archive (https://www.ncbi.nlm.nih.gov/sra) under accession number SRP141714.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The Natural Sciences and Engineering Research Council of Canada Bioconversion Network (NETGP350246-07), BioFuelNet and a Canada Research Chair to V.J.J.M supported this research. Damien Biot-Pelletier and Dominic Pinel were supported by a graduate scholarship from the Fonds de Recherche du Québec—Nature et Technologies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Population sequencing and alignment metrics. Table S2. Amplicon sequencing and alignment metrics for genotyping of R57 backcrossing isolates. Table S3. List of primers used for production of Ion Torrent sequencing libraries.
Additional file 2.
List of single nucleotide polymorphisms detected by whole population sequencing.
Additional file 3: Figure S1.
Evolutionary trajectories for all non-silent mutations identified by population genome sequencing at 6 time points. Mutations arose either in the MATα (left) or MATa (right) haploid populations. On the vertical axis are the names of the mutations, giving the closest gene, coordinates with respect to that gene and the nature of the nucleotide substitution. On the horizontal axis are each of the six evolutionary time points (UV, R1, R2, R3, R4, R5) and the mean allele frequency change (M). Frequencies of the mutant alleles are represented by shades of green. Mean allele frequency changes are represented in shades of red (M < 1, declining frequency) or blue (M > 1, increasing frequency). Hierarchical clustering of individual evolutionary trajectories is represented by dendrograms on the left. Mutations were assigned to groups of mutations (a1-5, α1-4) on the basis of this clustering. Mutations present in highly tolerant mutant R57 are highlighted in bold.
Additional file 4: Figure S2.
Evolutionary trajectories and apparent selection of all mutation hotspots identified by population sequencing. Mutations arose either in the MATα (left) or MATa (right) as indicated immediately to the left of each mutation. On the vertical axis are the names of the mutations, giving the closest gene, coordinates with respect to that gene and the nature of the nucleotide substitution. On the horizontal axis are each of the six evolutionary time points (UV, R1, R2, R3, R4, R5) and the mean allele frequency change (M). Frequencies of the mutant alleles are represented by shades of green. Mean allele frequency changes are represented in shades of red (M < 1, declining frequency) or blue (M > 1, increasing frequency). Mutations linked by connectors and marked with an asterisk indicate pairs with significantly similar initial frequency (binomial test, p > 0.05).
Additional file 5:
Supporting methods. Detailed methods are provided for our SNP calling methodology, structural study of Gdh1p, determination of mating type and ploidy by PCR, and modeling of SSL tolerance by multiple linear regression.
Additional file 6: Figure S3.
Backcrossing of R57 with wild type cells generates strains presenting a wide spectrum of fitness in SSL. Growth in the presence and absence of SSL is reported for R57, various wild type cell types and 86 F2 isolates from backcrossing of R57 and CEN.PK113-1A. Error bars represent plus or minus one standard deviation. The dashed line is a visual reference for the level achieved by the wildtype.
Additional file 7: Figure S4.
Genotyping of second generation segregants from backcrossing of R57 and wild type yeast suggest a model of SNP contributions to the SSL tolerance phenotype. Haploid (A) and diploid (B) isolates are scored in green for the genotypes indicated at the bottom. Growth in 85% SSL is scored in shades of blue on the right. Each row represents a single strain. Contribution to the phenotype of the indicated genotypes was inferred by multiple linear regression, yielding coefficients represented at the top in shades of red (diminishes fitness) to blue (increases fitness). Modeling of genetic interactions was attempted and the resulting coefficients are represented as circles at the top of the heatmaps. Growth in SSL predicted by the linear model is reported in shades of blue in the rightmost column, showing the level of agreement between the model and the data.
Additional file 8: Figure S5.
Single mutations are not sufficient to detect an increase in SSL tolerance in diploid cells. Area under the growth curve in the presence and absence of SSL for heterozygous (A) and homozygous (B) single diploid mutants is reported. Error bars represent plus or minus one standard deviation. The dashed line is a visual reference for the level achieved by the wildtype.
Additional file 9: Figure S6.
Reversion of nrg1 and gsh1 mutations leads to loss of the SSL tolerance phenotype in haploid single mutants. Area under the growth curve for nrg1 and gsh1 double mutants, haploid (nrg1 gsh1) and diploid (nrg1/nrg1 gsh1/GSH1) is also reported. Error bars represent plus or minus one standard deviation. The dashed line is a visual reference for the level achieved by the wildtype.
Additional file 10: Figure S7.
Growth in the presence and absence of SSL of single and double revertants identifies mutations contributing to the SSL tolerance phenotype. Area under the growth curve in the presence and absence of SSL is reported for (A) single haploid mutants, (B) single revertant derivatives of R57 and (C) revertant derivatives of R57 SGO1 gdh1-2/2, wild type for the indicated genes. Error bars represent plus or minus one standard deviation. The dashed line is a visual reference for the level achieved by the wildtype.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Biot-Pelletier, D., Pinel, D., Larue, K. et al. Determinants of selection in yeast evolved by genome shuffling. Biotechnol Biofuels 11, 282 (2018). https://doi.org/10.1186/s13068-018-1283-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-018-1283-9